Done
- Have spent much of my time this week running models using pydra-ml on hbn dataset and running models using regression (L1,L2,elastic net) and classification (decision tree, adaboost, random forest) to predict CGAS Score (continuous measure of general cognitive functioning) and diagnosis, respectively. A simple demographic model (features: sex, age) is not successful at predicting either of these measures, although there is a significant relationship between diagnosis and sex. Language tasks do reasonably well at predicting whether a participant has been diagnosed with a disorder or not (compared to physiologic function for example). It also seems, tentatively, that language tasks do better at predicting certain disorders, notably autism, however, I haven’t done significant testing yet.
- Technical challenges: I am doing the preprocessing step before I run pydra-ml, there were some challenges in passing
ColumnTransformer to the pipeline separately for both numeric and categorical data. I also would like to incorporate another step in pydra-ml that allows for model comparison (features + classifiers). Currently, one filename is passed in (one set of features) + multiple classifiers and model comparison is done on one set of features.
- Tried UMap in supervised mode to identify unique patterns in phenotypic data
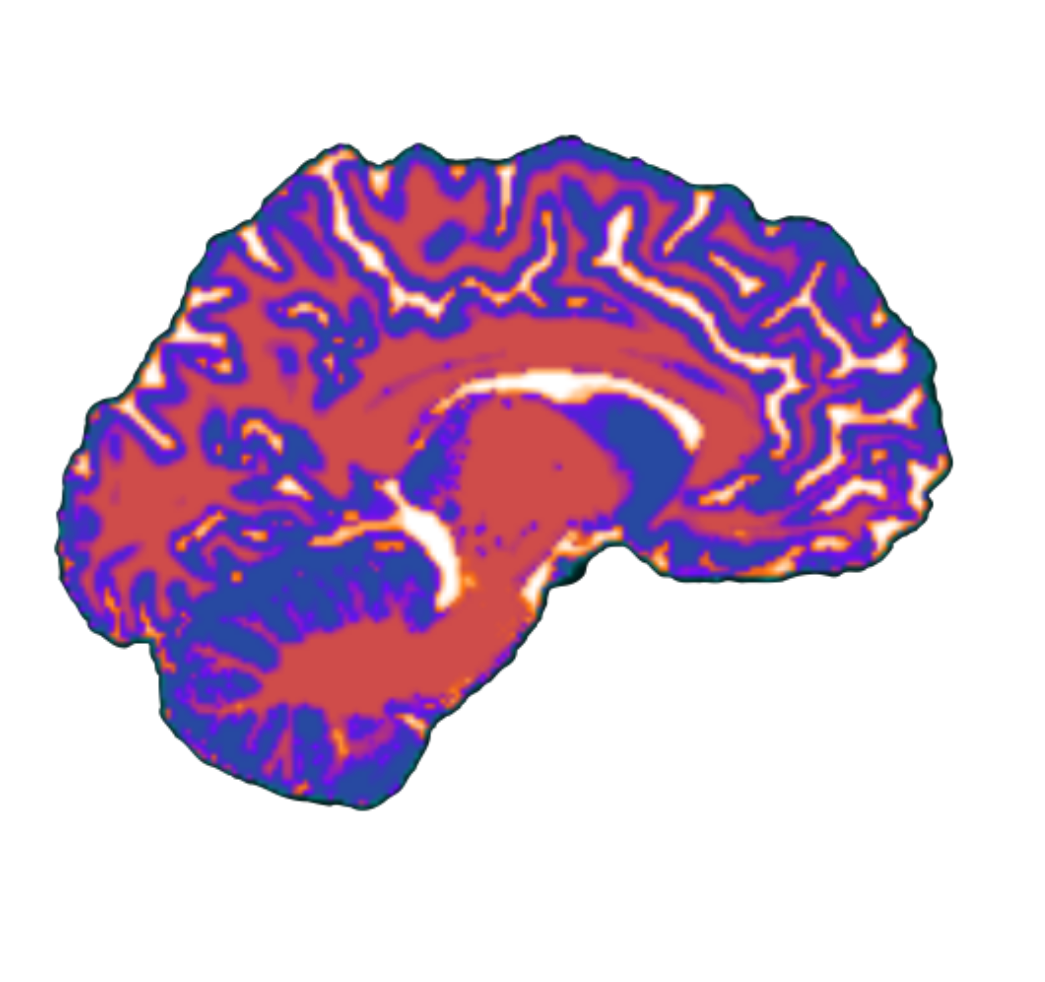
- Ironed out some bugs in
SUITPy for isolation and segmentation and tested it on a small subset of the HBN (across developmental stages). Each participant takes ~ 25 minutes and adds < 1GB of storage. Have yet to extend this analysis to >10 participants of HBN, will need to talk to Dorota/Hoda about space allocation on OpenMind, perhaps will do batch analysis.
- Interviewed UROP candidates for two positions, recruited one student from Wellesley to work on research methods, and have to decide between three great MIT candidates for a computational position.
To Do
- questions to answer: what is the reliability in the KSADS assessment across child and parent measures? meant to get to this question during the week but didn’t have time.
- Perform VBM to measure local changes in structural abnormalities, following methods steps implemented in this paper - correcting for brain size etc.
Questions
- Is there anything that I should do in advance of the CHOP meeting on Monday?
- What is the protocol for storing new analyses (
SUITPy output) on >2000 participants? I was planning on analyzing n=30 from each subgroup of HBN (adhd, autism, control, mood, anxiety, intellectual etc) as a first step, but ultimately I will want to analyze the broader dataset and will need to run SUITPy on each participant.