Done
- Wrote a predictive modeling pipeline using scikit-learn tools (and repurposing code I had written for previous projects). Worked reasonably well at doing basic model predictions on the HBN dataset. However, I explored pydra-ml, realized it was a lot better than what I had written, and decided to transition to using some of its functionality. My pipeline, for example, did not allow for model comparison as elegantly as pydra-ml. I was having difficulty doing preprocessing (using scikit-learn’s ColumnTransformer) within the framework and ended up trying to debug pydra-ml. This then led me down many hours of exploring (and learning) how to navigate pydra (not wasted time - I learned a lot).
- I also spent a lot of time on feature engineering this week, which I always forget (along with data cleaning, transformation etc.) takes more time than anticipated.
 .
.
- I have yet to incorporate jupyter notebooks into quarto framework (which I believe is possible) so I can’t share any of the initial prediction results here. Some of the questions I’ve addressed:
- Within HBN assessment battery, which tests are most useful? (how does this vary across disorders?)
- can further subtyping of individuals be done based on patterns in behavioral/cognitive assessments? (especially for 200 people who were not diagnosed due to an incomplete evaluation)
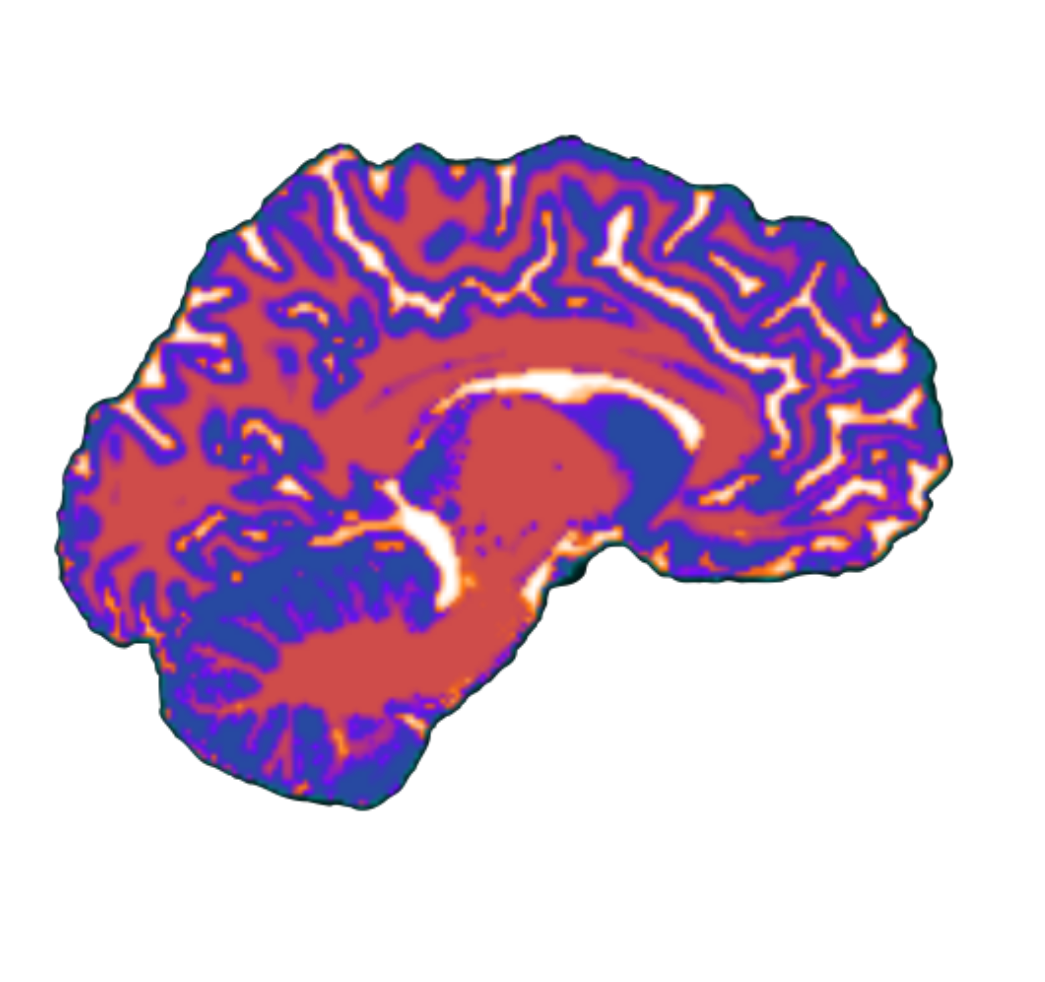
- I tested python implementation of SUIT on subset (n=10) of hbn participants (T1w + T2). There are still a few bugs to be ironed out in the python library (that don’t exist in matlab) - I will touch base with Joern about this in the next few days if I don’t figure it out myself. As we discussed in our last meeting, there are also deep learning approaches that are comparable (if not better) at isolating and segmenting the cerebellum. I haven’t investigated these approaches much but it might be necessary if we are analyzing cerebella of >3000 participants, and if SUIT is not reliable.
- Wrote an ad for two research assistants and circulated broadly (including UROP), have yet to receive any applications so likely that I will need to cast the net more widely.
To Do
- Try UMap in supervised mode to identify unique patterns in phenotypic data
- what is the reliability in the KSADS assessment across child and parent measures?
- Perform VBM to measure local changes in structural abnormalities, following methods steps implemented in this paper - correcting for brain size etc.
Questions
- Reached out to folks at HBN about getting LORIS access but haven’t heard from them yet. I’m analyzing the data from Openmind which I believe is up-to-date. I have access to all of the phenotypic data and I’ve been able to analyze some of the anatomical data that’s on OpenMind so I don’t think I’m missing anything, but would be nice to have full access so that I can stay current with any changes or updates.
- I’ve scheduled a meeting for next Wednesday to discuss some of the results from HBN and to talk about the CHOP project before our meeting with the Philadelphia group.
 .
.